The Chen Lab at NUS and GIS
Bacterial pathogenesis and genomics
Solutions for infectious diseases, antibiotic resistance, and synthetic biology
The lab has closed
From 2010-2022, the Chen Lab worked on bacterial pathogenesis and genomics at the National University of Singapore and the Genome Institute of Singapore. As of January 2023, the PI is working in a different industry, and the lab is officially closed.
This website remains as a partial record of the accomplishments of the former members of the lab, and may receive some updates as remaining papers are published.
Key strains cited in our publications have been preserved. If you are looking for these, or for other scientific correspondence related to the publications of the Chen lab, you may contact Swaine Chen at swainechen_AT_gmail_DOT_com.
Our Vision
We are working to integrate modern sequencing and computational methods into the daily discovery process of microbiologists.
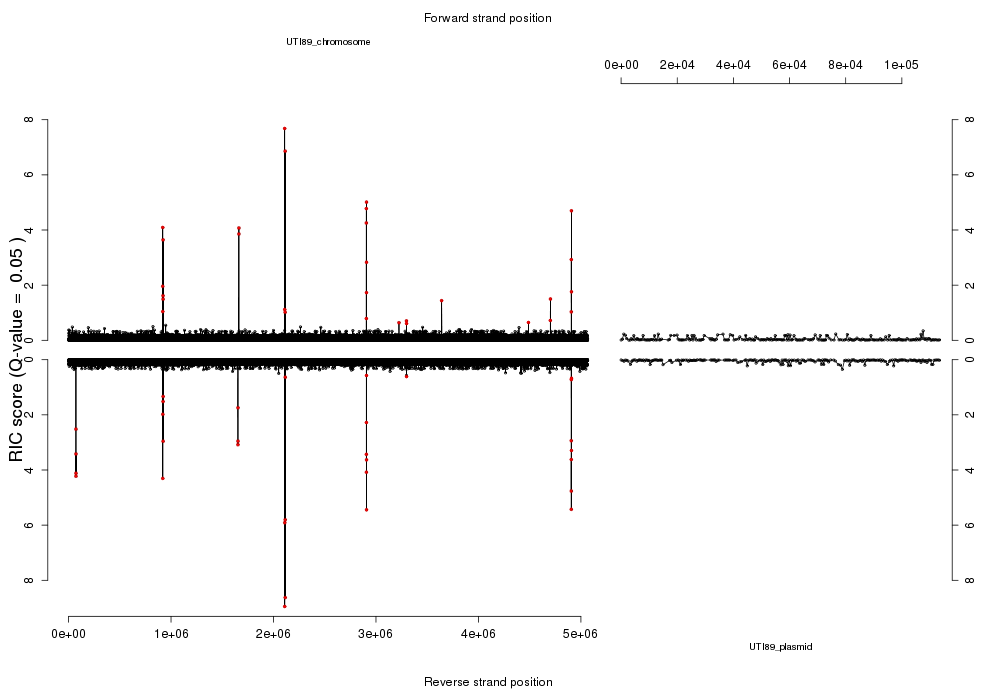
Urinary tract infections caused by Escherichia coli are a major unmet disease need which we study in detail. The UTI model system also serves as a testbed for developing sequencing-powered methods that could be applied to other infectious diseases.
Research in the lab falls into three major themes:
In vivo
Molecular interactions underly disease.
We need to understand these molecular details in order to treat or prevent infections. Urinary tract infections and foodborne Group B Streptococcus disease are our main experimental models.
In vitro
Genetic techniques are needed to understand bacteria causing disease today.
Traditional genetic methods have been developed for specific lab-adapted strains. We are forging new technologies to manipulate strains that are cuasing dieasese in humans today. These techniques are always more general and sometimes more powerful as well.
In silico
How can we increase the pace of discovery?
Not all infectious diseases have good model systems. However, we can collect DNA sequence from nearly any organism. Can we leverage computational techniques, using only DNA sequences, to gain mechanistic insights into infectious diseases? This would truly increase our ability to understand and combat disease.
Learn about our Research
Experimentally, we work on two main systems: urinary tract infections caused by Escherichia coli and invasive foodborne infections caused by Streptococcus agalactiae (Group B Streptococcus). We also have some explicitly computational projects which are exploring how bacteria, and their ability to cause disease, evolves. We also have interests in the spread of antibiotic resistance genes and synthetic biology. Click below for more details!